This web page was created as an assignment for Genetics 677, an undergraduate course at UW-Madison.
Gene Ontology and Phenotypes
The outward human phenotype of Hutchinson-Gilford progeria syndrome has been discussed on previous pages of this website. However, the cellular phenotype that leads to the outward appearance and complications of HGPS have not been covered in as much death and may be the most important aspect at identifying this disease and possibly finding a viable treatment for it. The data for this page was assembled from geneontology.org, Phenobank, and WormBase. These sites give insight into the role LMNA or its homologs play in an organism's biological process, cellular component, and molecular function, as well as the displayed phenotype.
The gene ontology for LMNA was found using geneontology.org, which is a database that attempts to standardize the vocabulary surrounding genes, their products, and the variety of roles it might play within an organism. By doing so this database tries to facilitate uniformity across several database using different tools making it easier for researchers to find information about a given gene. A query ofLMNA for humans yielded six unique accession terms covering the three ontologies (given in parentheses): muscle development (biological process), lamin filament (cellular component), nuclear envelope (cellular component), perinuclear region of cytoplasm (cellular component), protein binding (molecular function), and structural molecular activity (molecular function). Furthermore ontologies looking at biological processes and molecular function were the least specific for LMNA associations, since such processes or very complex and involve a number of different genes and their associated product. The most specific results was for the cellular components of lamin filaments, which would be expected as the gene codes for lamins A and C, and the perinuclear region. These results coupled with an association with the nuclear envelope agree with research done with this gene, as patients with HGPS have abnormalities in the nuclear envelope structure. These results returned by GO are evidenced through citing journal articles that had previously conducted experiments to determine function or process of LMNA or from direct assays done regarding this gene. Knowing the ontology of LMNA gives an idea into the phenotype that should be displayed.

The genomic environment of lmn-1, the homolog used to study phenotype of LMNA. Further genomic position data can be found here.
Phenotypes for the gene were investigated using Phenobank and WormBase (click on these links for LMNA search results). These databases use results from experiments done on the LMNA homologs in C. elegans. WormBase gives an intensive and in depth look into the phenotype and ontolgy of the gene from direct experimentation and RNAi. In fact the ontology from this database is much more specific.
Brief description of Lmn-1 (homolog of LMNA): It is the only nuclear lamin in C. elegans and is part of the intermediate filament superfamily. Essential for nuclear localization of Emerin protein during embryonic development and binds to mitotic chromosomes and histone 2A.
Expression: It is found is localized around the nuclear envelope during all developmental stages and all regions where the Ce-lamin antibodies (used for staining) penetrated. Using Ce-lamin-GFP fusion protein, lmn-1 was found in nuclear periphery during all developmental stages, with areas within this space more highly fluorescent. More specifically, lmn-1 was found to localize underneath the inner-membrane of the nucleus. All of these findings corroborate what researchers phenotypes found in HGPS patients, mainly the integral role LMNA plays in cellular development and the structural integrity of the nuclear envelope.
Phenotypes: A variety of phenotypes were found using RNAi or direct experimentation. A large portion of these caused pre-natal lethality or sterility. These phenotypes could be true in humans but would not be documented since the affect child would not come to term and those who are born do not live long enough to test for sterility. There were phenotypes found in the C. elegans experiments that are observed in living HGPS patients including: shorter life span, abnormal aging, and pronuclear abnormalities.
Below is a movie recording of an RNAi experiment done to observe the effects if lmn-1 is silenced. As you can see the shape of the nucleus is distorted indicating problems with the structure of the nuclear envelope and membrane. Similar cellular phenotype are seen in individuals with HGPS.
| lmn-1.mov | |
| File Size: | 3891 kb |
| File Type: | mov |
Chemical Genetics
Using PubChem's Bioassay search engine for the LMNA gene, three results were returned, all related to HGPS research. PubChem is a database that includes chemical experiments to examines the effect small molecules can have on the activity of specific proteins. Each of the assays investigated splicing modulators, which could be expected as HGPS patients have an activation of a cryptic splice site that leads to disfunctional lamins A and C. Looking at the second result, high throughput techniques were used to test over 190,000 compounds and found 27 active compounds. The other two experiments looked at about 200 compounds and found no active compounds. The result page gives an overview of the experiments carried out, the protocol used, and links to the results found.
AID: 1487
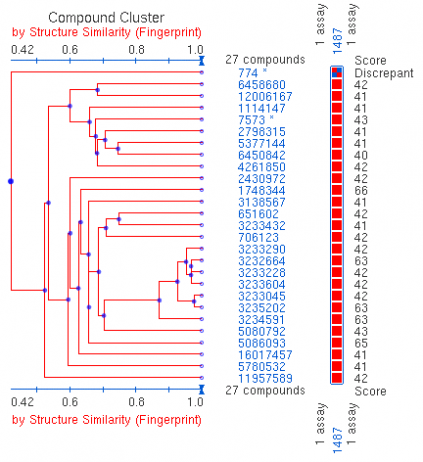
Fig. 1 This figure represents the cluster family for the qHTS assay of lamin A splicing. The scores to the right indicate the similarity between the compound isolated and the target site being tested for.
Compound SID CID Score Result Potency
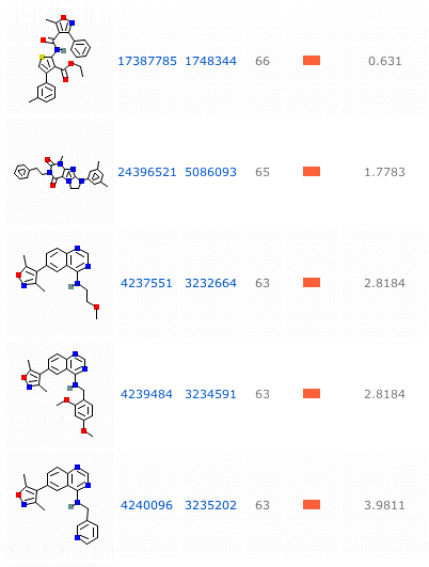
Fig. 2 The results in this figure show the highest rated active compounds found during the chemical tests looking at splicing modulators in isoform 3 of LMNA. The figure displays the structures of the small molecules as well as their scores. This figure shows the five highest scored compounds. These were the only compounds with a score above 43. The potency of these compounds refers to the affinity for that compound to bind to the target site.
References
All data found on this page was collected from: geneontology.org, Phenobank, WormBase, PubChem.
Peter St. Andre [email protected] Last updated: 4/11/09.
gen677.weebly.com
